Use of a four-tiered graph to parse the factors leading to phenotypic clustering in bacteria: a case study based on samples from the Aletsch Glacier
Miroslav Svercel, Manuela Filippini, Nicolas Perony, Valentina Rossetti and Homayoun C. Bagheri
PLOS ONE (2013)
Abstract
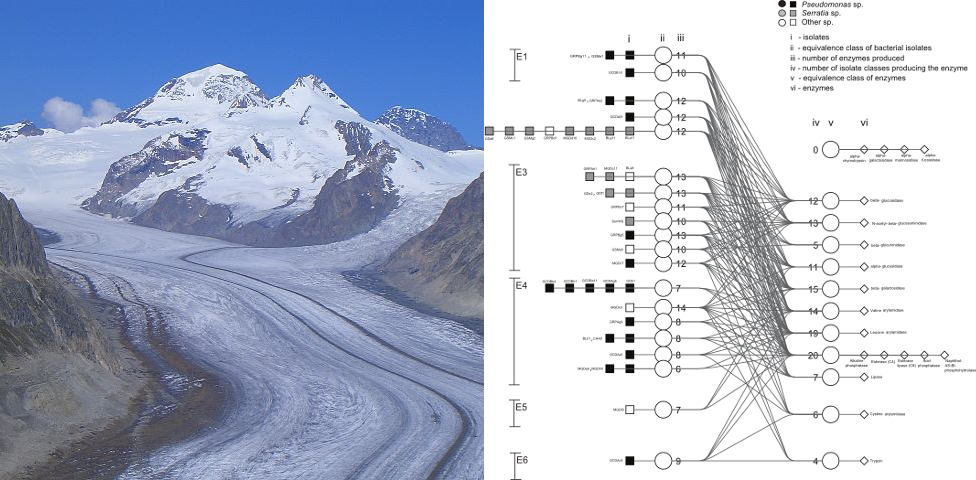
An understanding of bacterial diversity and evolution in any environment requires knowledge of phenotypic diversity. In this study, the underlying factors leading to phenotypic clustering were analyzed and interpreted using a novel approach based on a four-tiered graph. Bacterial isolates were organized into equivalence classes based on their phenotypic profile. Likewise, phenotypes were organized in equivalence classes based on the bacteria that manifest them. The linking of these equivalence classes in a four-tiered graph allowed for a quick visual identification of the phenotypic measurements leading to the clustering patterns deduced from principal component analyses. For evaluation of the method, we investigated phenotypic variation in enzyme production and carbon assimilation of members of the genera Pseudomonas and Serratia, isolated from the Aletsch Glacier in Switzerland. The analysis indicates that the genera isolated produce at least six common enzymes and can exploit a wide range of carbon resources, though some specialist species within the pseudomonads were also observed. We further found that pairwise distances between enzyme profiles strongly correlate with distances based on carbon profiles. However, phenotypic distances weakly correlate with phylogenetic distances. The method developed in this study facilitates a more comprehensive understanding of phenotypic clustering than what would be deduced from principal component analysis alone.